Corn Disease Detector (CDD)
Early plant disease detector model built using neural networks. Saturdays AI Quito 2021.
Welcome to Corn Disease Detector (CDD) Main Page

Authors: Juan Zurita, Sebastián Ayala Ruano, Ximena Celi, Gilberto Rodríguez.
Infectious diseases are a major threat for many crops of high importance for the food security of many regions of the world. Each year, around 60% of farmers in Ecuador have reported pre-harvest losses due to infectious agents such as fungi and bacteria, in crops like banana, cacao, and potato. Outbreaks of infectious plant diseases could endanger the country’s economy and cause many people to lose their jobs. This project is an effort to develop an early plant disease detector as a proof of concept, using publicly available datasets of maize. In the future, we plan to expand this project to cover other important farming Ecuadorian species.
Dataset
The image dataset splited in training, validation, and test used to build our model is available at this link.
About our model
Our model is a classifier based on convolutional neural networks, trained to recognize two types of maize infectious diseases: Common rust of corn and Northern corn leaf blight. The available datasets for maize are too small to be used in deep neural networks. To solve this problem we used a Transfer Learning strategy and reused some layers from a ResNet50 neural network trained on the ImageNet dataset, from torchvision.models.resnet50. In this way, general image patterns are identified by pre-trained layers, and we trained additional layers of the network to identify healthy and diseased corn images. This network architecture has shown very promising results in previous studies of computer vision for agriculture.
The main library used to build our model was Pytorch and we trained it using Google Cloud GPUs through Google Colab.
Model Performance
The loss and accuracy of training and validation datasets are shown below:
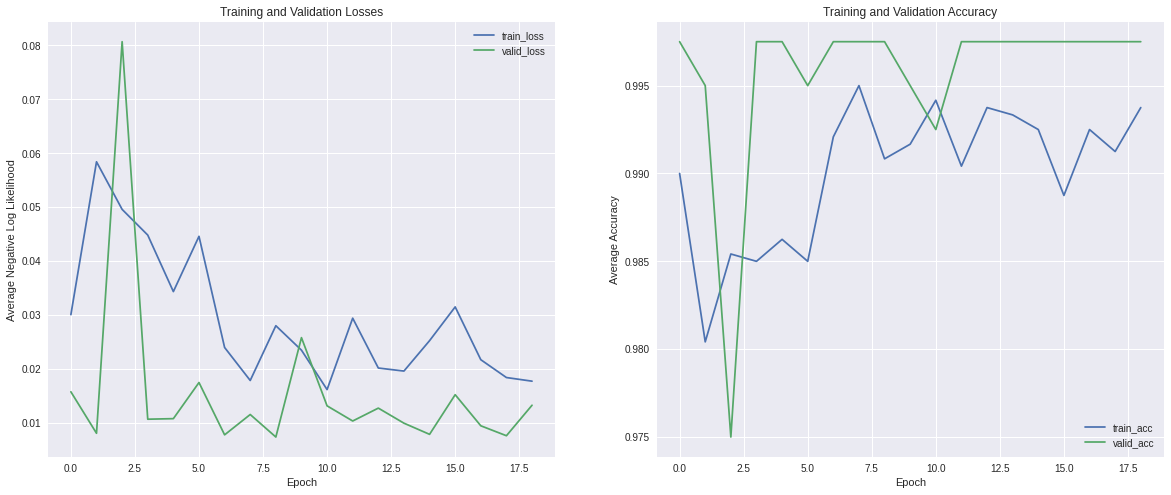
In both datasets, the loss decreased and the accuracy increased through training epochs, as we expected. Also, validation loss values were less than the training counterpart and accuracy had the opposite behavior, which means that is not likely that our model is overfitting.
To gain insight into the generalization capacity of our model on new data, we calculated some performance measures on the test dataset, including the confusion matrix, accuracy, precision, recall, F1 score, and Matthews correlation coefficient. The summary of these metrics is presented below:
Confusion Matrix
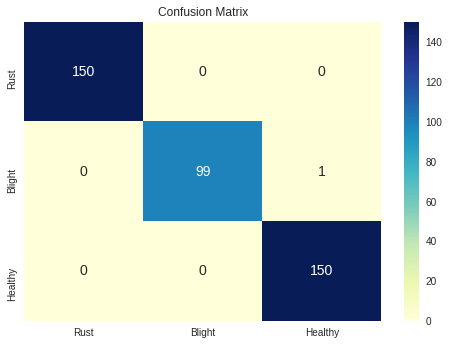
| Precision | Recall | Accuracy | F1-score | |
|---|---|---|---|---|
| Common rust of corn | 0.99 | 1 | 1 | 1 |
| Northern corn leaf blight | 1 | 0.98 | 0.99 | 0.99 |
| Healthy | 0.99 | 1 | 1 | 1 |
Matthews correlation coefficient: 0.99
Run the Streamlit app locally
In case you just want to run the model, you can access our web application with this link.
Also, you can run the model locally using our GitHub repository. First, you should clone the main branch of our repo to your machine, decompress the contents of the zip file, and change your working directory:
$ git clone https://github.com/corndiseasedetector/corndiseasedetector.github.io
$ unzip corndiseasedetector.github.io-main.zip
$ cd corndiseasedetector.github.io
We created a requirements file which has all the Python libraries you will need to run the app locally. We highly recommend installing the libraries using pip in a new Python virtual environment (version 3.8.11) using pyenv version manager (there were some errors using conda). The creation of a separate virtual environment helps to prevent version conflicts with your OS and other personal projects. If you are not familiar with pyenv, this blog is a good introduction to this useful tool. The following command can be used to create a new virtual environment with pyenv:
$ pyenv install -v 3.8.11
Before installing the libraries with pip, please check out that your working Python version is the one you created with pyenv:
$ python -V
Then, install all Python libraries and dependencies of the requirements.txt file:
$ pip install -r requirements.txt
When the installation is finished, run this command:
$ streamlit run https://github.com/corndiseasedetector/corndiseasedetector.github.io/blob/main/webapp.py
Now, you can test the app locally with your images. The app will look like this when active through Streamlit.
Run the Streamlit app in the cloud
If you do not want to install the dependencies to run our model locally, you can access our model hosted as a web service in this link.
Replicate our work
If you wish to replicate our work, you could make a copy of the jupyter notebook we used to build, train, and test our model, to your Google Drive and run it with Google Colab. Also, you can run the notebook on your computer if you have GPUs, or make the proper changes to work with CPUs.
Support or Contact
If you have any request for trouble using our model, please create a pull request in the GitHub repository, or you can contact us at our email ai2021grupo7@gmail.com.